This has to be a quick one, but I wanted to go on the record is noting my frustration at the current concern that Ebola might "mutate" into something far worse, like a pathogen that is efficiently transmitted by aerosol. For example, Michael Osterholm wrote in the New York Times yesterday, "The second possibility is one that virologists are loath to discuss openly but are definitely considering in private: that an Ebola virus could mutate to become transmissible through the air." I heard Morning Edition host David Greene ask WHO Director Margaret Chan last week, "Is this virus mutating in a way that could be very dangerous, that could make it spread faster?"
I agree, Ebola Virus becoming more easily transmitted by casual contact would be a 'nightmare scenario.' However, what we need to worry about is not mutation per se, but selection! Yes, the virus is mutating. It's a thing that viruses do. Ebola Virus is a Filovirus. It is composed of a single strand of negative-sense RNA. Like other viruses, and particularly RNA viruses, it is prone to high mutation rates. This is exacerbated by the fact that RNA polymerases lack the ability to correct mistakes. So mutations happen fast and they don't get cleaned up. Viruses also have very short generation times and can produce prodigious copies of themselves. This means that there is lots of raw material on which selection can act, because variation is the foundation of selection. Add to that heritability, which pretty much goes without saying since we are talking about the raw material of genetic information here, and differential transmission success and voilà, selection!
And virulence certainly responds to selection. There is a large literature on experimental evolution of virulence. See for example the many citations at the linked to Ebert's (1998) review in Science here. There are lots of different specific factors that can favor the evolution of greater or lesser virulence and this is where theoretical biology can come in and make some sense of things. Steve Frank wrote a terrific review paper in 1996, available on his website, that describes many different models for the evolution of virulence. Two interesting regularities in the the evolution of virulence may be relevant to the current outbreak of EVD in West Africa. The first comes from a model developed by van Baalen & Sabelis (1995). Noting that there is an inherent trade-off between transmissibility of a pathogen and the extent of disease-induced mortality that it causes (a virus that makes more copies of itself is more likely to be transmitted but more viral copies means the host is sicker and might die), they demonstrate that when the relative transmissibility of a pathogen declines, its virulence will increase. They present a marginal value theorem solution for optimal virulence, which we can represent graphically in the figure below. Equilibrium virulence occurs where a line, rooted at the origin, is tangent to the curve relating transmissibility to disease-induced mortality. When the curve is shifted down, the equilibrium mortality increases. EVD is a zoonosis and it's reasonable to think that when it makes the episodic jump into human populations, it is leaving the reservoir species the biology of which it is adapted to and entering a novel species to which it is not adapted. Transmission efficiency very plausibly would decrease in such a case and we would expect higher virulence.
The second generality that may be of interest for EVD is discussed by Paul Ewald in his book on the evolution of infectious disease and (1998) paper. Ewald notes that when pathogens are released of the constraint between transmissibility and mortality -- that is, when being really sick (or even dead) does not necessarily detract from transmission of the pathogen -- then virulence can increase largely without bound. Ewald uses the difference in virulence between waterborne and directly-transmitted pathogens to demonstrate this effect. At first glance, this seems to contradict the van Baalen & Sabelis model, but it doesn't really. The constraint is represented by the curve in the above figure. When that constraint is released, the downward-sloping curve becomes a straight line (or maybe even an upward-sloping curve) and transmissibility continues to increase with mortality. There is no intermediate optimum, as predicted by the MVT, so virulence increases to the point where host mortality is very high.
A hemorrhagic fever, EVD is highly transmissible in the secretions (i.e., blood, vomit, stool) of infected people. Because these fluids can be voluminous and because so many of the cases in any EVD outbreak are healthcare workers, family members, and attendants to the ill, we might imagine that the constraints between transmissibility and disease-induced mortality on the Ebola Virus could be released, at least early in an outbreak. As behavior changes over the course of an outbreak -- both because of public health interventions and other autochthonous adaptations to the disease conditions -- these constraints become reinforced and selection for high-virulence strains is reduced.
These are some theoretically-informed speculations about the relevance of selection on virulence in the context of EVD. The reality is that while the theoretical models are often supported by experimental evidence, the devil is always in the details, as noted by Ebert & Bull (2003). One thing is certain, however. We will not make progress in our understanding of this horrifying and rapidly changing epidemic if all we are worried about is the virus mutating.
Selection is overwhelmingly the most powerful force shaping evolution. The selective regimes that pathogens face are affected by the physical and biotic environments in which pathogens are embedded. Critically, they are also shaped by host behavior. In the case of the current West African epidemic of EVD, the host behavior in question is that of many millions of people at risk, their governments, aid organizations, and the global community. People have a enormous potential to shape the selective regime that will, in turn, shape the pathogen that will infect future victims. This is what we need to be worrying about, not whether the virus will mutate. It saddens and frustrates me that we live in a country where evolution is so profoundly misunderstood that even our most esteemed, and otherwise outstanding sources of information and opinion don't understand the way nature works and the way that human agency can change its workings for our benefit or detriment.

![log[Y(t)]/t](http://monkeysuncle.stanford.edu/wp-content/plugins/latex/cache/tex_380b1cea9aadb2220e7ff6b252ec1556.gif)
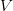
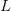
![\log[Y(t)]/t](http://monkeysuncle.stanford.edu/wp-content/plugins/latex/cache/tex_60ed99c4631e6be2f173613ae73e67f9.gif)